M.S. Candidate: Ümit Sude Böler
Program: Bioinformatics
Date: 01.09.2025 / 11:00
Place: A-212
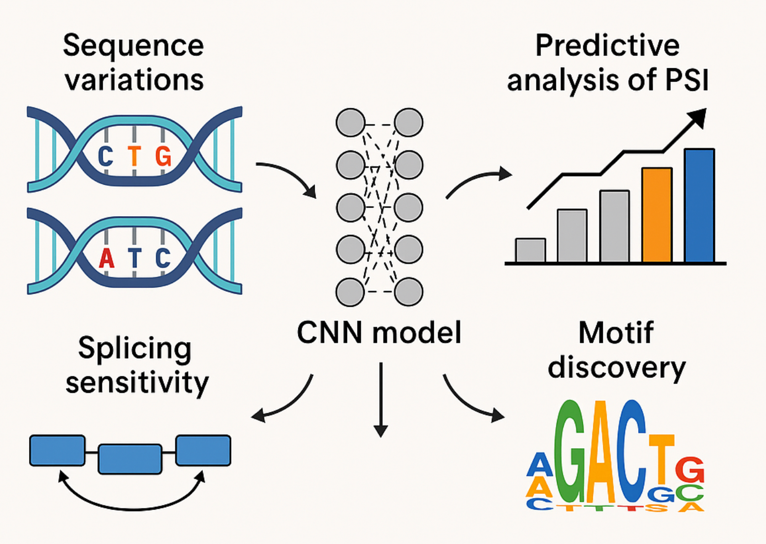
Abstract: Alternative splicing is essential for the expansion of transcriptomic complexity, with microexons rep- resenting some of the most functionally critical and tightly regulated splicing events in neural develop- ment. Under differential SRRM4 expression, this thesis examines the effects of engineered sequence variants in the downstream intron, microexon, and upstream intron on Percent Spliced In (PSI) values. Using a synthetic microexon library from a Massively Parallel Splicing Assays (MaPSy), a custom convolutional neural network (CNN) model was developed to predict splicing outcomes under four SRRM4 response conditions (GFP, LOW, MID, HIGH). The model was trained on one-hot encoded sequences and auxiliary metadata, achieving strong predictive performance across conditions. In order to improve the biological interpretability, DeepLIFT-based feature attribution and TF-MoDISco-lite were implemented to extract regulatory sequence motifs that contribute to PSI predictions. The dis- covered motifs were then annotated using TOMTOM against the CISBP-RNA database, providing insights into potential co-regulatory elements associated with SRRM4-mediated splicing modulation. This study offers a computational framework for deciphering the cis-regulatory logic of microexon in- clusion and highlights how integrative modeling can advance our understanding of splicing regulation in neurodevelopmental contexts.