M.S. Candidate: Hüseyin Hilmi Kılınç
Program: Bioinformatics
Date: 07.01.2025 / 10:00
Place: A-212
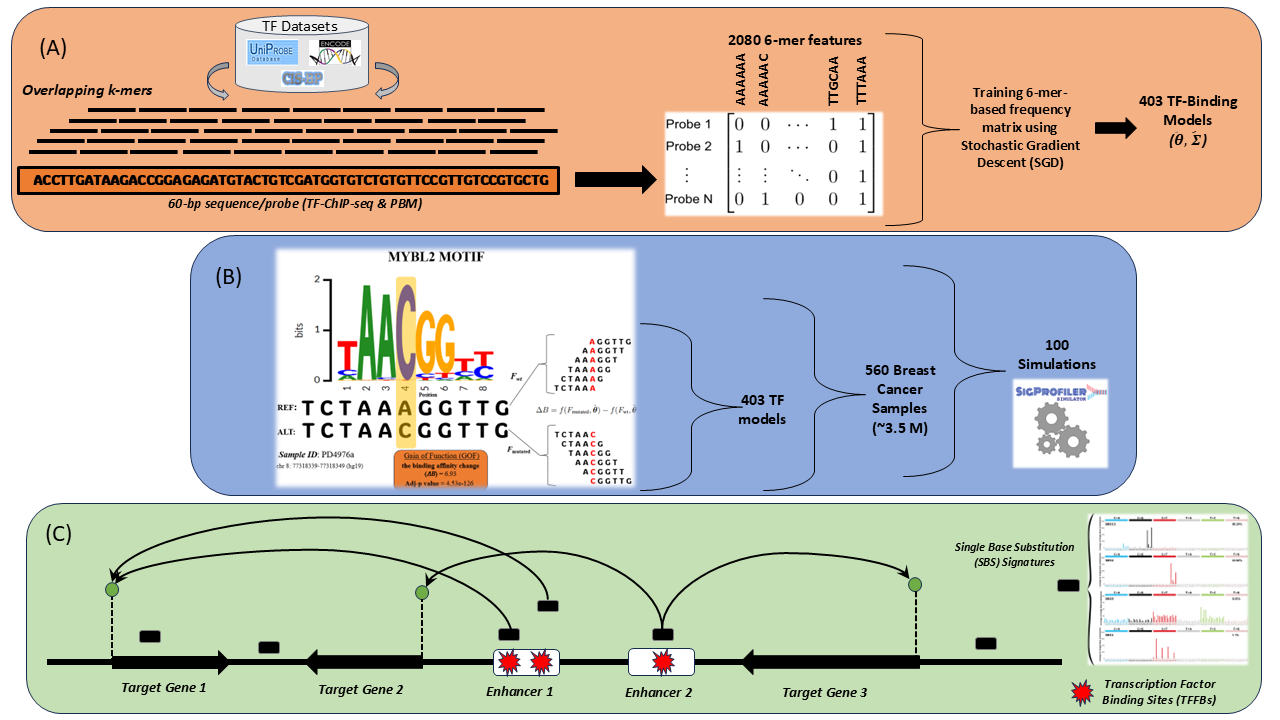
Abstract: Somatic mutations, particularly in non-coding regions, can perturb transcription factor (TF)-DNA interactions, influencing gene regulatory networks and contributing to cancer development. Thus, they require further comprehensive analysis to explore their effect on this regulatory interplay. In this study, we developed an in silico pipeline that assesses the impact of these somatic mutations on TF-binding affinities. In our computational framework, we built k-mer-based linear regression models for 403 human TFs trained on high-throughput in vivo (ChIP-seq) and in vitro protein-binding microarray (PBM) datasets employing the stochastic gradient descent (SGD) algorithm. Applying our predictive TF models to somatic mutations of 560 breast cancer samples from a large cohort study, we quantitatively predicted changes in TF binding affinities, assigning them gain of function (GOF) or loss of function (LOF) effects. We further explored the perturbations of TF family-wide trends due to certain mutational signatures imprinted by distinct mutagenic processes. These TF binding changes were integrated with putative enhancer-target gene maps derived from the activity-by-contact (ABC) model, linking potential dysregulations affecting oncogenes and tumor suppressor genes in breast cancer. We also conducted subtype-specific analyses to explore distinct patterns of TF perturbations across breast cancer molecular subtypes. Finally, the 560 breast cancer datasets were simulated 100 times using the SigProfilerSimulator tool, which constructs a tailored null hypothesis for statistical analysis of our observed patterns. Consequently, our pipeline can prioritize somatic mutations with potential regulatory effects in cancer genomics, advancing our understanding of non-coding mutations and their role in cancer genomics.