Ph.D. Candidate: Umut Çınar
Program: Information Systems
Date: 19.06.2023 / 15:45
Place: B-116
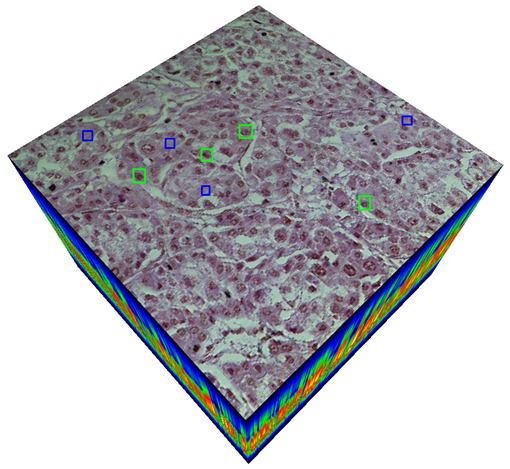
Abstract: In this study, we introduce a new method for classifying Hepatocellular Carcinoma (HCC) using a hyperspectral imaging system (HSI) integrated with a light microscope. We developed a custom imaging system that captures 270 bands of hyperspectral images from healthy and cancerous tissue samples with HCC diagnosis taken from a liver microarray slide. To build an accurate classification model, we utilized Convolutional Neural Networks (CNNs) with 3D convolutions (3D-CNN). These convolutions incorporate both spectral and spatial features within the hyperspectral cube to train a robust classifier. By leveraging 3D convolutions, we can collect distinctive features automatically during CNN training without requiring manual feature engineering on hyperspectral data. Our proposed method is compact and can be applied effectively in medical HSI applications. Additionally, we addressed the class imbalance problem in the dataset by utilizing the focal loss function as the CNN cost function. This function emphasizes hard examples to learn and prevents overfitting caused by the lack of inter-class balancing. Our empirical results indicate that hyperspectral data outperforms RGB data in liver cancer tissue classification, and increased spectral resolution leads to higher classification accuracy. Furthermore, we found that spectral and spatial features are both critical for training an accurate classifier for cancer tissue classification.