M.S. Candidate: Ramal Hüseynov
Program: Bioinformatics
Date: 13.01.2026 / 11:00
Place: A-212
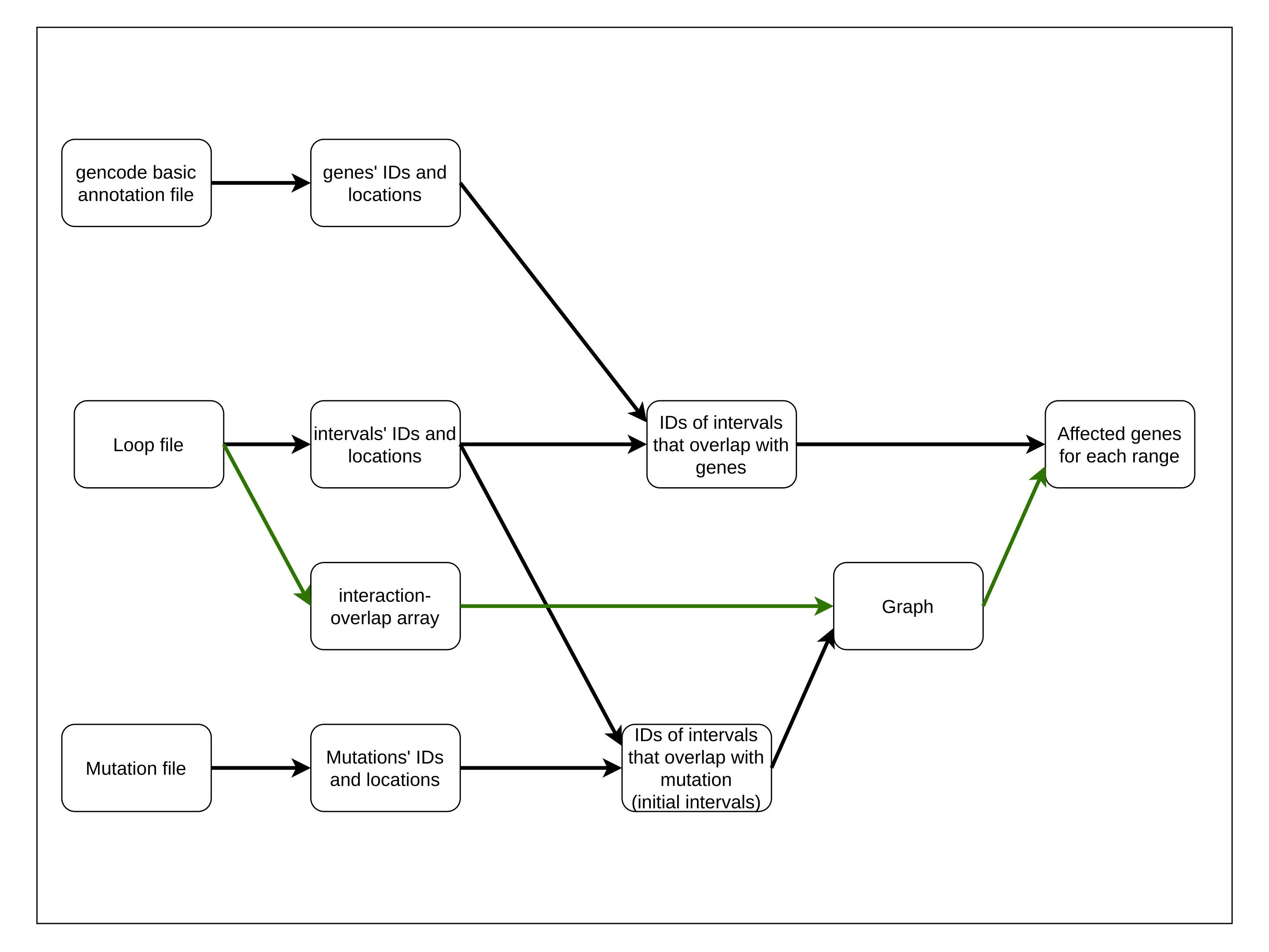
Abstract: Somatic mutations drive the transformation of normal cells into cancer. However,distinguishing driver mutations, which confer a selective growth advantage, from the vast background of neutral passenger mutations remains a critical challenge. To address this, we introduce a novel graph-based framework that constructs mutation-centric networks by leveraging longrange genomic interaction data. Our method models genomic intervals as nodes and their long-range interactions or overlaps as edges. Starting from a ’seed’ mutation, the graph expands iteratively, finding overlaps and interacting intervals to capture both local and distal genomic context. This architecture allows us to quantify a mutation’s topological influence, identify complex structural patterns (such as graph cycles), and assess proximity to known driver genes across variable ranges. Furthermore, this approach naturally generates embeddings for individual mutations, enabling the clustering of samples based on mutation profile similarity. Ultimately, by providing a comprehensive, interaction-aware view of the genomic landscape, our framework facilitates more accurate driver identification and improved patient stratification for personalized treatment.