PhD. Candidate: Aslı Boyraz
Program: Medical Informatics
Date: 18.11.2022 / 11:30
Place: A212
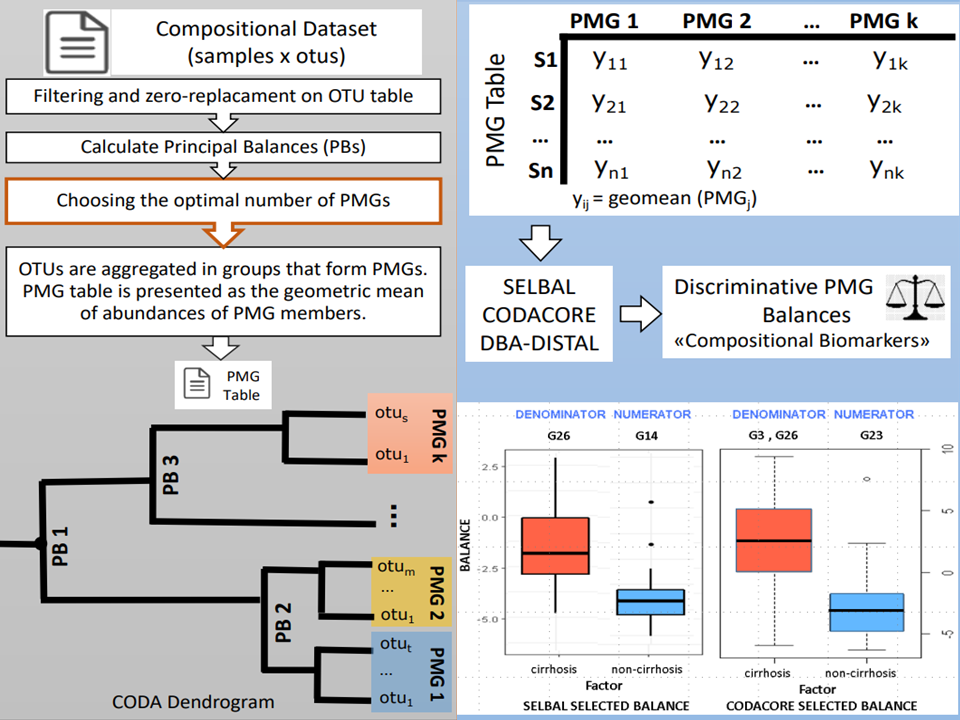
Abstract: The microorganisms present in the human body play a crucial role in maintaining human health, and the environmental microbiome influences the human microbiome. Advanced understanding of the human microbiome and indoor microbiota is the first step towards understanding the potential relationships between health and microbiome. Next Generation Sequencing (NGS) enables identification and study of a large number of microorganisms in a short time. With the identification of a large number of microorganisms, the studies for the understanding of their role in the environment and human health have become important. This thesis examines the production and the properties of microbiome data and statistical challenges of microbiome analysis. First, we give a brief history of the various methods of analysing microbiome data. We are mainly concerned with performing microbiome analysis using compositional approaches. The proposed procedures were illustrated with the data from 16S rRNA amplicon sequencing but those also apply for microbiome shotgun metagenomics. This dissertation describes the basics of compositional data (CoDa) analysis introducing log-ratio methodology. The first part of this thesis deals with the problem of establishing relationship based on the microbial features annotated with taxonomic information, where a compositional alternative to phylogenetic grouping of microbiome data (Principal Microbial Groups - PMGs) is proposed to enable working with low-level microbial features (OTUs or ASVs). The usefulness of the proposed procedure is illustrated on a Cirrhosis dataset to search for biomarker candidates. The second part of the thesis focuses on the microbial transmission and PMGs are aimed to investigate any hint to track microbial transmission. An experiment that was conducted at Erciyes University Hospital for this purpose, and swab samples were gathered from the Intense Care Unit (ICU) to construct microbiome profiles. Microbial transmission is carried out between objects, so it is expected that resulting microbiome profiles of samples should have similar microbial structure. In this case, not taxonomic changes but OTU/ASV abundance changes between samples need to be investigated. PMGs procedure were applied to microbial transmission dataset in order to analyze the contagion. PMGs provide a valid grouping for OTUs alternative to taxon grouping using CoDa approach and it offers the possibility of working with coarse group of OTUs, which are not present in a phylogenetic tree in microbiome analysis.