Ph.D. Candidate: Melike Çağlayan
Program: Medical Informatics
Date: 18.01.2023 / 10:00
Place: A212
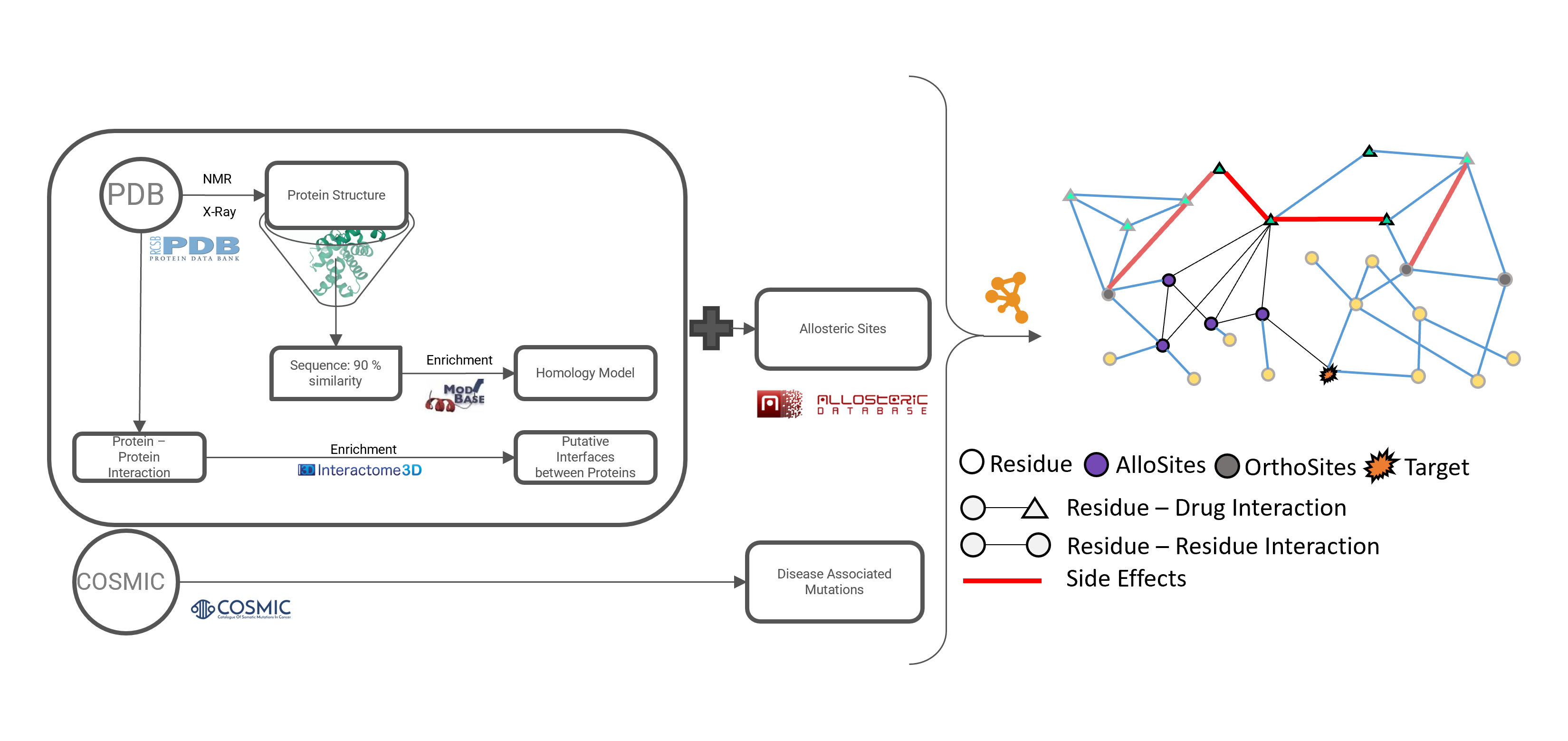
Abstract: Proteins play a central role in many biological processes and their function is regulated by various mechanisms including allosteric regulation. Allostery refers to the regulation of protein activity by molecules or ions binding at sites other than the active site. Despite the importance of allostery, understanding the mechanisms of allosteric regulation and identifying allosteric pathways in protein structures has been a challenging task due to the complexity and diversity of protein structures. In this research, we proposed a novel structure-based network approach for examining allosteric protein regions. We represented each protein and protein complex structure as a residue-residue interaction network and used this representation to identify allosteric pathways connecting two distant sites. We also developed a computational method to predict the presence and location of allosteric regions in protein structures and examined the effect of these regions on protein structure in terms of whether they originate from the core or surface of the protein. To validate our method, we applied it to a dataset of 14 proteins of known structure that are regulated allosterically and used statistical metrics to assess the accuracy of the results. Our analysis showed that the residues obtained by our method and their effect on the allosteric pathway were highly accurate. In the future, we plan to investigate the potential connection between allosteric sites and drug-binding sites, as well as the impact of mutations on allosteric pathways and their overlap with disease-causing mutations and phosphorylation sites.